Community resource
CHAMBER is a free community resource. It can be readily accessed and used by all stakeholders, under the terms of a CC BY-SA 4.0 licence.

Interactive
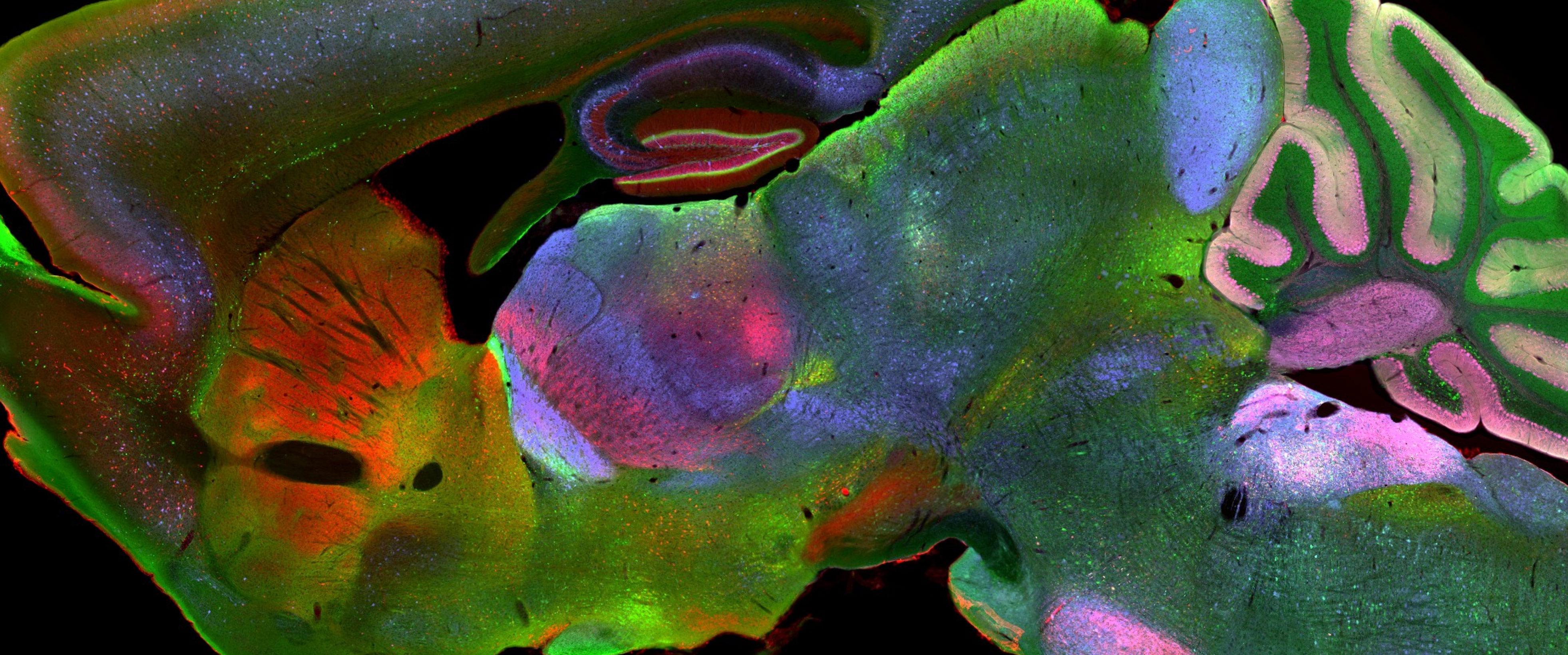
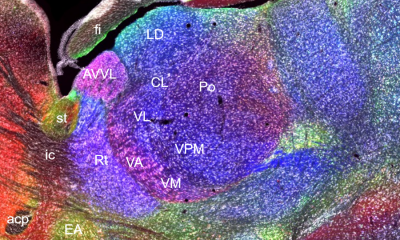
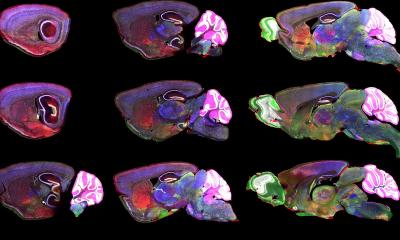
CHAMBER’s images have submicron resolution, are annotated for regions of interest and stereotaxic landmarks, offer a ‘Google Maps-like’ navigation experience, come with detailed experiment metadata, and can be downloaded for further use.

Team
The CHAMBER team members are Kouichi C. Nakamura, Ben Micklem, Giulio Spagnol, Naomi Berry, Andrew Sharott, and Peter J. Magill. The advantages of using chemoarchitectonic data to delineate functionally-specialized brain regions are highlighted by the team’s study of the motor thalamus (see Nakamura et al. 2014). Not so coincidentally, the term thalamus originates from the Greek word thalamos (θάλαμος), which means “[inner] chamber".