Using iViewer
CHAMBER uses the open-source OMERO platform with OMERO.iviewer, developed by The Open Microscopy Environment.
You can navigate the images in CHAMBER quite intuitively.
-
Mouse wheels or two-finger pinching, depending on devices, provide zoom in and out.
-
Dragging the window invokes scrolling.
-
If you hold Shift key while dragging, you can rotate the image.
![]()
In addition to the basic manipulations above, at the top of the iviewer window, you'll find these tool buttons as above.
The buttons at the top left are for zooming out, zooming in, original view, and pixel size view ![]() .
.
You can also specify the magnification by giving a percentage ![]() .
.
At the top right, there is the indicator for the XY coordinates with the plus button to toggle intensity querying (currently unstable) ![]() ,
,
the arrow button to reset the image rotation ![]() ,
,
and the double arrow button for full screen view ![]() .
.
We highly recommend to try the full screen view ![]() , especially on a large screen!
, especially on a large screen!
By default, when you enter CHAMBER using the links above, you'll find the side panel on the right. Click the Setting tab for customizing how the images are displayed.
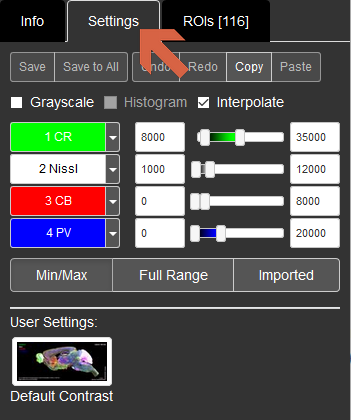
Save, Save to all: These buttons are disabled for public users.
Undo, Redo, Copy, Paste: You can Undo or Redo your action on Settings. Copy will copy the current settings to the clipboard and you can Paste it to other images.
Grayscale: If you tick this, only the current channel will be shown in grayscale.
Interpolate: This is checked by default. At above 1:1 pixel zoom, the display is interpolated so that you don't see pixelation.
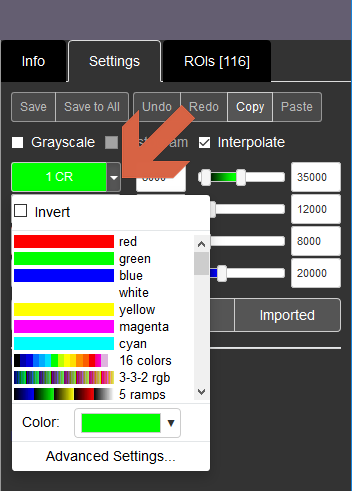
Channel names: The channel name is written over its pseudo colour on the left of the sliders. Hovering your cursor over the truncated text will make the full channel name appear. The colour-filled channel names work as toggle buttons to turn on and off each channel. By clicking the down arrows to their right, you can change the channel colors or lookup tables (LUTs). You can also invert the values of each LUTs.
Sliders and boxes: used to set the display range of each channel via black and white points.
Min/Max: Set the display range based on the minimum and maximum values in present each channel.
Full Range: sets the white and black points for each channel to the minimum and maximum values possible in the images.
Imported: uses the black and white point values set in the file before it was imported into OMERO.
User Settings: We provide a Default Contrast preset so you can always return to the original display settings (channel colours and display ranges) after making adjustments.
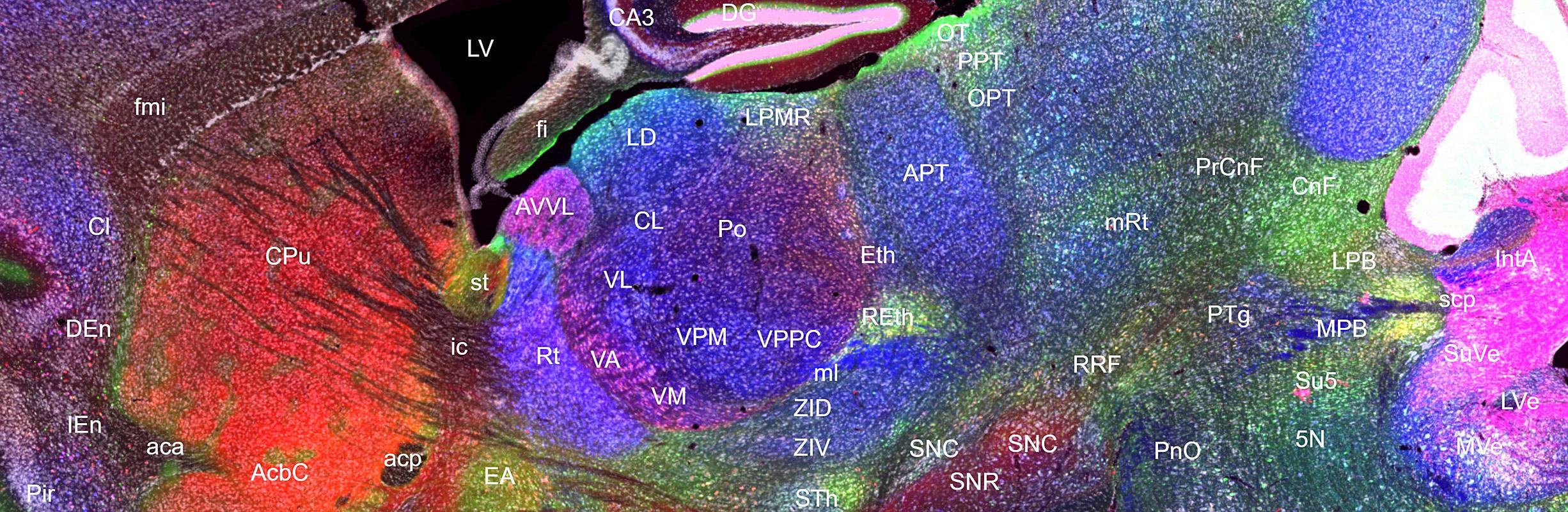
1. Currently, you need to click the ROIs tab in the right side panel to show annotations (mostly the names of structures).
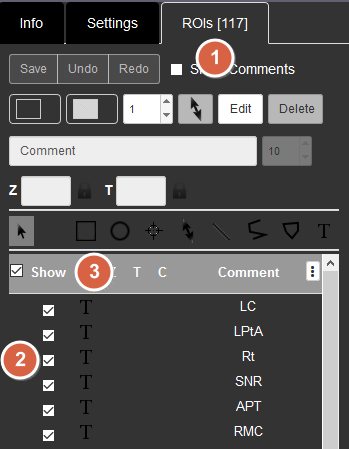
2. You can show and hide individual ROIs (region of interests) separately by ticking or unticking the boxes on the left side of the list.
3. Or show and hide all the ROIs at once by ticking/unticking the box next to Show at the top.
4. When you click an ROI on an image, that ROI will be shown on the list on the right.
Structural annotations and abbreviations used for kms024v1v4 dataset is a subset of those used in The Mouse Brain in Stereotaxic Coordinates Third Edition by Keith B.J. Franklin and George Paxinos (Academic Press, 2007) with a few exceptions.
A full list of abbreviations will be added soon.
The ROIs include two vertical yellow lines in case of parasagittal planes and they represent the bregma and interaural lines in terms of anterior-posterior axis.
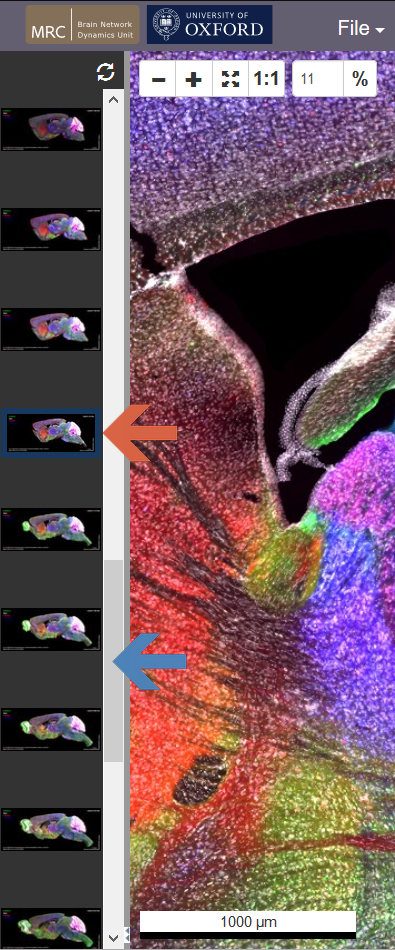
When you access the CHAMBER using the links above, you'll see the thumbnails of other images in the same dataset on the left. The current image is indicated by the blue outline (red arrow). Use the vertical slider (blue arrow) and click on one of the other thumbnails to load a new image.
Close to the bottom of the screen, at the borders between the left- or right-side panels and the main panel with the tissue image, you'll see a small button with two horizontal triangles.

You can hide/show the side panel by clicking this button (red circle).
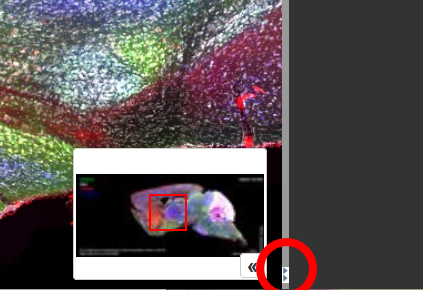
Image database
Method 1 (Screen Resolution)
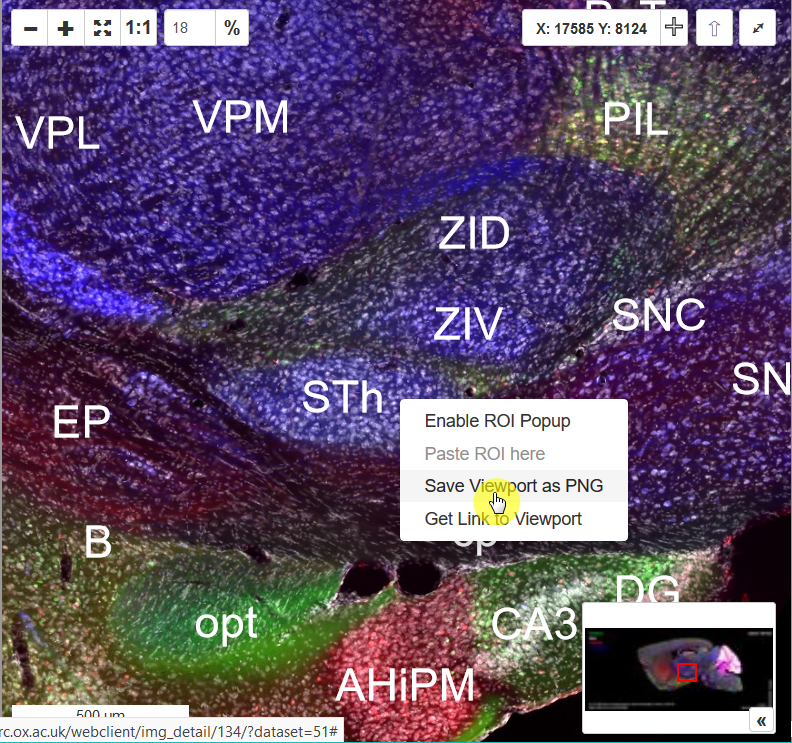
Right-click the navigation window, and you'll see a context menu with an option "Save Viewpoint as PNG." Select this and save the image as it appears on the screen at the screen resolution.
Method 2 (Full Resolution)
By default, when you enter the CHAMBER using the links above, you'll find the side panel on the right.
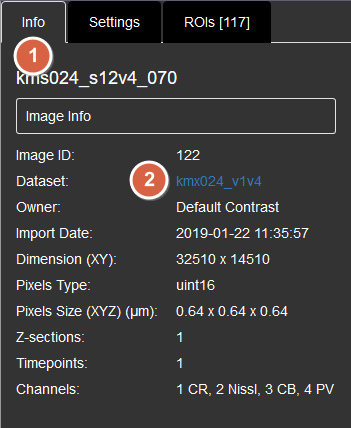
1. Click the Info tab.
2. Then click the blue URL link at "Dataset" property to jump to the web client page, where all the images are shown.
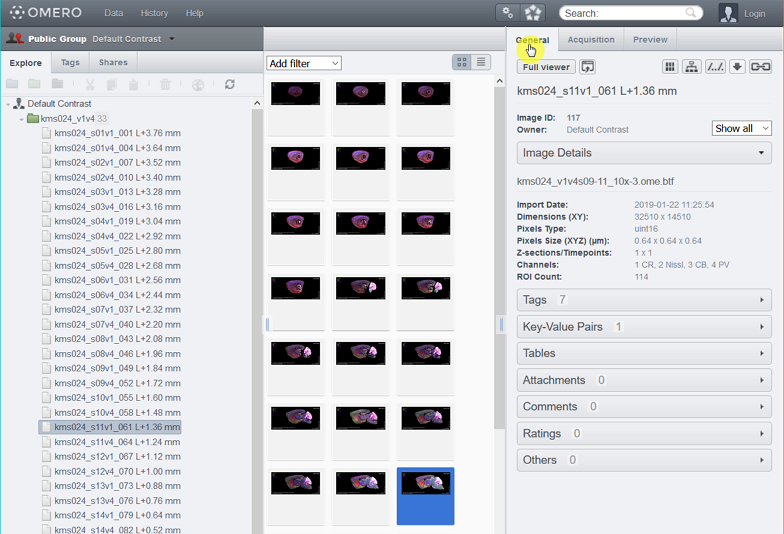
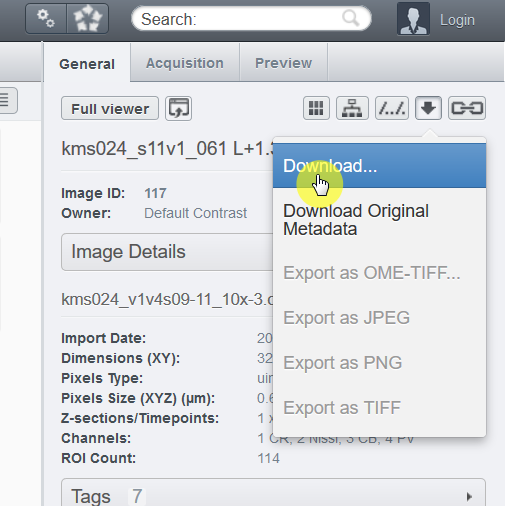
3. From the web client page, find the download icon at the General tab on the right side of the window.
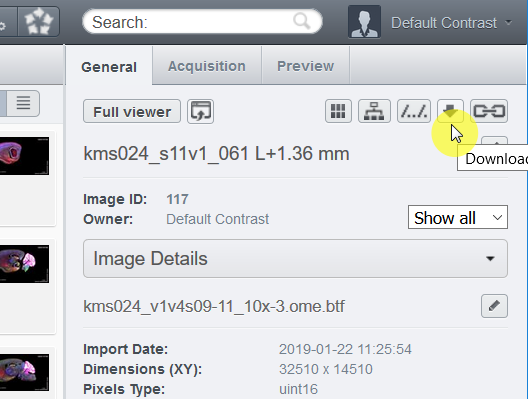
4. Choose "Download..." and you can download the whole image at the full resolution as the OME.TIFF format.
CHAMBER images come with complete metadata, such as dilution of antibodies, filters used for each channel, objective lens, and camera.
By default, when you enter the CHAMBER using the links above, you'll find the side panel on the right.
1. Click the Info tab.
2. Then click the blue URL link at "Dataset" property to jump to the web client page, where all the images are shown.


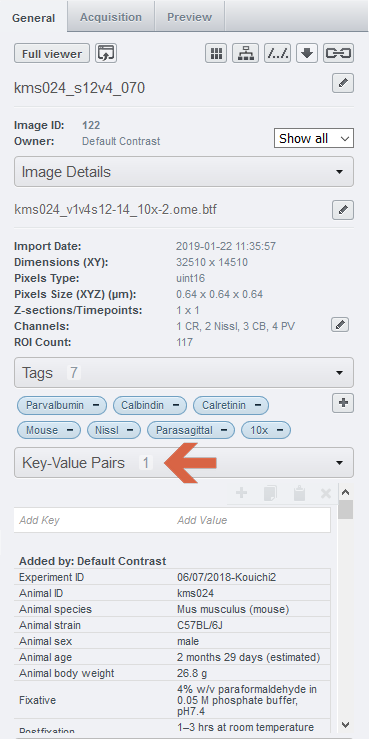
3. The right side panel of the web client page contains metadata for each image or dataset. On the General tab, the Key-Value Pairs hold detailed information about protocols of immunohistochemstry etc.
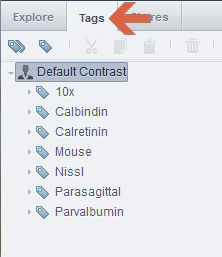
4. The Tags tab on the left side panel allows you to search for images and datasets with specific tags.
We want to maximise the value of CHAMBER to stakeholders, and we welcome feedback from current and prospective users.
We will add more proteins to the image databases. We anticipate organising workshops with key stakeholders to help us prioritize markers/antibodies. If you have a suggestion of protein markers/antibodies to add to CHAMBER, please write to Dr Kouichi C. Nakamura (kouichi.nakamura@pharm.ox.ac.uk).
CHAMBER is a curated resource, which helps us ensure that the experimental work, data and metadata are of the highest quality. If you have an image dataset that you would like to share via the CHAMBER platform (the dataset does not have to be a completed atlas), please write to Dr Kouichi C. Nakamura (kouichi.nakamura@pharm.ox.ac.uk). We are happy to give advice on the acquisition and post-processing of images.
We plan to create an online discussion forum to encourage stakeholder feedback on any aspect of CHAMBER. We are already active members of the online forums of Allen Brain Map and OMERO.
We plan to expand the scope and utility of the CHAMBER database, as follows:
1. Expand the image datasets for adult mouse brain, adding more proteins in both parasagittal and coronal sections (initially focusing on markers of axon terminals and major neurotransmitter systems, and based on community feedback).
2. Create image datasets for adult monkey (Macaca mulatta) brain, in a likewise manner.
3. Increase usability through the addition of web-based search functions, novel parcellation of regions, accurate stereotaxic referencing, and alignment with/to the Allen Mouse Common Coordinate Framework.
If you have any suggestions, please write to Dr Kouichi C. Nakamura (kouichi.nakamura@pharm.ox.ac.uk).
CHAMBER is a free resource, and registration is not required to use it. The datasets stored in CHAMBER are published under the Creative Commons Attribution Share-Alike 4.0 International (CC BY-SA 4.0) license. The publisher and copyright holder is the University of Oxford. The CC BY-SA 4.0 license allows sharing and copying of CHAMBER data in any medium or format, and the adapting or processing of the data for any purpose, including commercially.
If you use any data obtained from CHAMBER, the license requires that you give appropriate credit, such as a citation if used in an academic publication (see ‘How do I cite CHAMBER?’), and indicate if changes have been made. If you distribute data or derived data from CHAMBER, the license requires that this is distributed under the same license.
If you use any data from CHAMBER for an academic publication, please cite CHAMBER accordingly. This is important to fulfill the ‘attribution’ element of the CC BY-SA 4.0 license, but also helps us evaluate the use of CHAMBER. Formatting styles for citations will vary, but should still contain the creators, year of publication, title, publisher and DOI:
Nakamura KC, Micklem BR, Berry NSM, Spagnol G, Sharott A, Magill PJ (2019) CHAMBER: Calcium-binding proteins (calretinin, calbindin, and parvalbumin) with Nissl staining on parasagittal sections of mouse brain [dataset] (University of Oxford) doi:10.5287/bodleian:8gYKJX7Ob
We monitor online visits to CHAMBER so that we can fulfil funder requirements for assessing impact. We do not record the last three digits of visitor IP addresses, which means visitors cannot be identified, but we can see their geographic locations. We also gather data about device type, operating system and screen resolution; this information helps us to improve the functionality and display of CHAMBER. The MRC BNDU hosts CHAMBER on internal servers, and these data are never transferred to a third party
Accurate empirical determination of different brain regions - as defined by their specialised inputs, outputs and constituent cell types - is critical for understanding brain function. Revealing brain ‘chemoarchitectonics’, that is, features emerging from distinct patterns of protein expression, offers clear opportunities for more accurately defining regions and cell types. However, images of brain tissue protein expression in the research literature can be difficult to interpret, partly because most publication formats preclude end users from readily navigating and interrogating the images for themselves. Many images are also placed behind paywalls, further reducing their utility. Thus, major benefits can be gained by improving the quality, usability, and accessibility of chemoarchitectonic data for delineating mammalian brain regions. The creation of CHAMBER, with its freely-accessible online archives of high-resolution images, presents a novel framework to achieve this. We anticipate that CHAMBER will fuel discovery, encourage data sharing, and provide important advances for the ‘3Rs’.