Analysis of hippocampal ensembles during Contextual Feeding and cNOR tasks
This dataset contains two jupyter notebooks and python scripts to run some exemplar analysis from the article 'Organising the coactivity structure of the hippocampus from robust to flexible memory'.
The jupyter notebooks provided are:
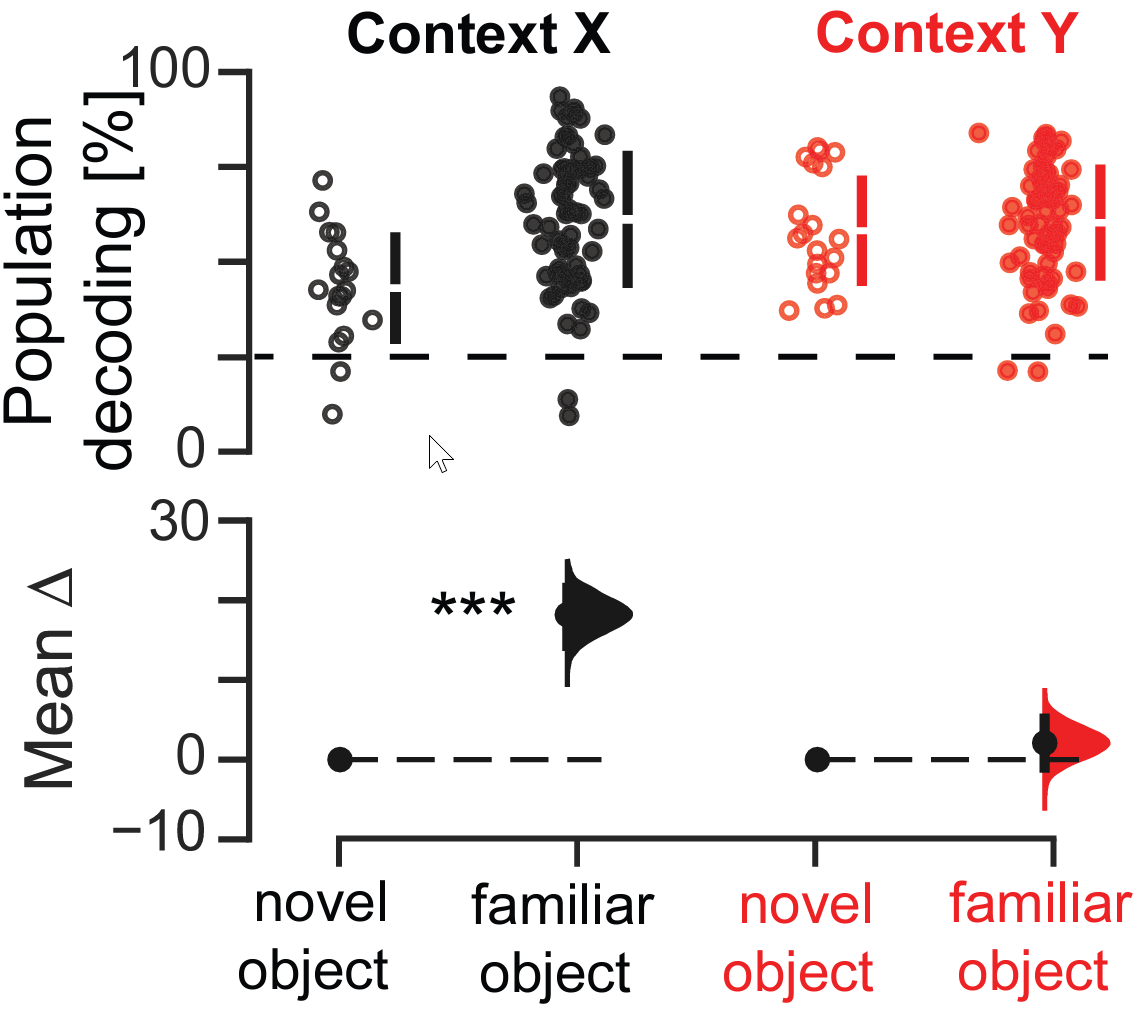
- 'cNOR_task.ipynb' which computes the object/location coding analyses shown in Figures 1 and 4
- 'coactivity_cond+cnor.ipynb' reproduces some of the coactivity analyses shown in Figures 1 and 4.
In the 'results' folder are stored processed data that are loaded throughout the notebooks.
The python script 'makeGraphbatch.py' computes coactivity graphs during active exploration times (theta-informed) from the spiking data. After downloading, ensure the spiking data folder is named 'data' and placed inside the root folder 'orgCoactHippo'. See script for more info.
Inside the 'recordings' folder, there are text files that list the recording days belonging to each task. That is: 'cond_ll145' and 'cond_ll149' lists the food-context conditioning days for each animal, while `cnor_x` and `cnor_y` list the cnor days in the two contexts, regardless of the animal's identity.
The python library 'util_func.py' data loading and processing functions used by the python script and notebooks. See script for more info.
'difference_estimation_plot.py' is a python library to produce estimation plots as in the article.
For the analysis in the original paper, this code was run in Python version 3.10. Execution of the code requires the following libraries:
matplotlib 3.7.1
matplotlib-inline 0.1.6
networkx 2.8.4
numpy 1.24.3
pandas 1.5.3
pandas-ods-reader 0.1.4
scikit-learn 1.2.2
scipy 1.10.1
seaborn 0.12.2
We welcome researchers wishing to reuse our data to contact the creators of datasets. If you are unfamiliar with analysing the type of data we are sharing, have questions about the acquisition methodology, need additional help understanding a file format, or are interested in collaborating with us, please get in touch via email. Our current members have email addresses on our main site. The corresponding author of an associated publication, or the first or last creator of the dataset are likely to be able to assist, but in case of uncertainty on who to contact, email Ben Micklem, Research Support Manager at the MRC BNDU.
Creative Commons Attribution-ShareAlike 4.0 International (CC BY-SA 4.0)
This is a human-readable summary of (and not a substitute for) the licence.
You are free to:
Share — copy and redistribute the material in any medium or format
Adapt — remix, transform, and build upon the material for any purpose, even commercially.
This licence is acceptable for Free Cultural Works. The licensor cannot revoke these freedoms as long as you follow the license terms. Under the following terms:
Attribution — You must give appropriate credit, provide a link to the license, and indicate if changes were made. You may do so in any reasonable manner, but not in any way that suggests the licensor endorses you or your use.
ShareAlike — If you remix, transform, or build upon the material, you must distribute your contributions under the same licence as the original.
No additional restrictions — You may not apply legal terms or technological measures that legally restrict others from doing anything the licence permits.