Behaviour and pupillometry in a bandit task
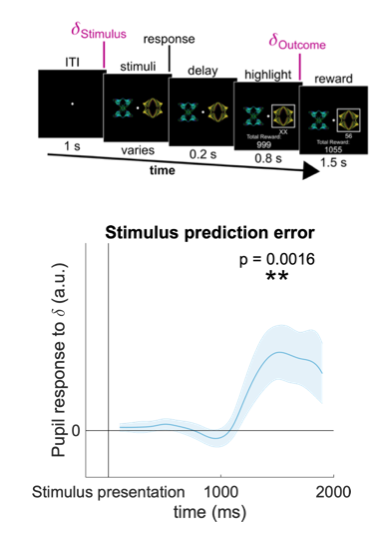
Our task consisted of sequences of 120 two-alternative forced choice trials. On each trial two stimuli were drawn from a set of four stimuli and shown to the participant, who had to choose one. Following the choice a numerical reward between 1 and 99 was displayed under the chosen stimulus. Then, the next trial began. Participants were instructed to try to maximize the total number of reward points throughout the experiment. The reward on each trial depended on the participant’s choice: each stimulus was associated with a specific reward distribution from which rewards were sampled. The four reward distributions associated with the four stimuli were approximately Gaussian and followed a two-by-two design: the mean of the Gaussian could be either high or low (60 or 40), and the standard deviation could be either large or small (20 or 5), resulting in four reward distributions in total ((60, 20), (60, 5), (40, 20) and (40,5)). We recorded choices and pupil dilation traces from 27 human subjects, which completed 4 di!erent sequences (blocks) of 120 trials each. The data they generated is formatted as a MATLAB table named "dt", with each row corresponding to a single trial, and each column (accessible by name) corresponding to a measured variable.
Columns:
- ID: unique participant identifier.
- Block: Block number
- trial: Trial number
- stim1: ID of stimulus on the left side of the screen.
- stim2: ID of stimulus on the right side of the screen.
- stim_chosen: ID of the chosen stimulus
- rt: reaction time in seconds.
- reward: reward in points.
- pupil_stimuli: pupil dilation time series, sampled at 10 Hz, % change relative to baseline, first sample time-locked to stimulus onset.
- pupil_reward: pupil dilation time series, sampled at 10 Hz, % change relative to baseline, first sample time-locked to reward onset
Stimulus ID corresponds to associated reward distribution (1: (60, 20), 2: (60, 5), 3: (40, 20), 4: (40, 5)). For a more detailed description of the task and pupil data
processing, see bioarxiv pre-print.
We welcome researchers wishing to reuse our data to contact the creators of datasets. If you are unfamiliar with analysing the type of data we are sharing, have questions about the acquisition methodology, need additional help understanding a file format, or are interested in collaborating with us, please get in touch via email. Our current members have email addresses on our main site. The corresponding author of an associated publication, or the first or last creator of the dataset are likely to be able to assist, but in case of uncertainty on who to contact, email Ben Micklem, Research Support Manager at the MRC BNDU.
Creative Commons Attribution-ShareAlike 4.0 International (CC BY-SA 4.0)
This is a human-readable summary of (and not a substitute for) the licence.
You are free to:
Share — copy and redistribute the material in any medium or format
Adapt — remix, transform, and build upon the material for any purpose, even commercially.
This licence is acceptable for Free Cultural Works. The licensor cannot revoke these freedoms as long as you follow the license terms. Under the following terms:
Attribution — You must give appropriate credit, provide a link to the license, and indicate if changes were made. You may do so in any reasonable manner, but not in any way that suggests the licensor endorses you or your use.
ShareAlike — If you remix, transform, or build upon the material, you must distribute your contributions under the same licence as the original.
No additional restrictions — You may not apply legal terms or technological measures that legally restrict others from doing anything the licence permits.