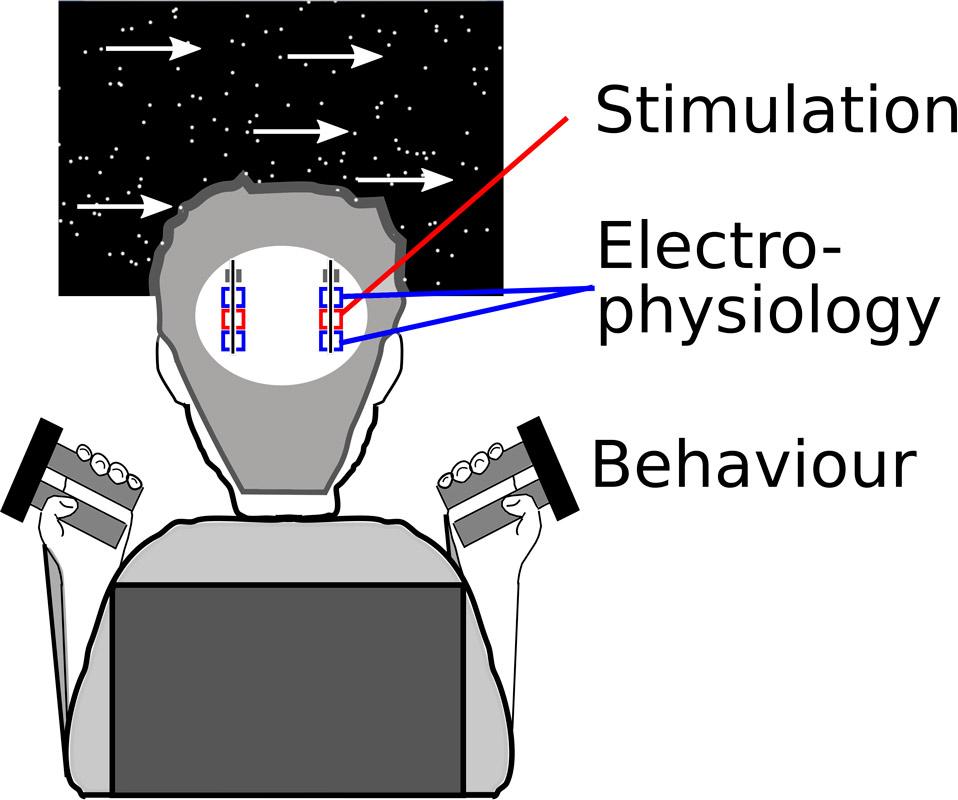
Subthalamic nucleus correlates of decision and movement speed.
This code analyses behavioural data from a group of 13 Parkinson patients and 15 healthy control participants performing a moving dots paradigm with Speed vs. Accuracy instructions. The main behavioural outcomes are reaction times, movement times, peak force and response accuracy (folder 1, used for figure 1 in the published article), as well as computational analysis of behaviour with hierarchical drift diffusion modelling (folder 2, used for figure 2). In patients local field potentials were recorded during the task and corresponding code is stored in folder 3-5 (figure 3). In 10 patients burst deep brain stimulation was applied during a second session. Its effects on behaviour and local field potentials are analysed with code from folder 6 and 7 (figures 4-6). The results have been published in a paper termed ‘Dynamic control of decision and movement speed in the human basal ganglia’ by Herz et al.
The code has been tested on a MacBook Pro, macOS Mojave 10.14.6. The required software is listed in each part of the analysis including where installation information can be found. Installation times vary between the different softwares. Run times of code is longest for Bayesian analysis (~ 5-10 minutes per analysis) and drift diffusion modelling (up to several hours). The remaining code in R and Matlab is much faster.
Example data is provided as HDDM.csv (example behavioural data of Parkinson’s disease (PD) patients), HDDM_HC.csv (example behavioural data of healthy control (HC) participants) and HDDM_LFP (combined example behavioural and local field potential data from patients). This is not the actual data from the study, and thus the results will not match the study results, but the structure is the same and therefore they allow testing all scripts if applicable. Note that for this data the log transform should be omitted, since the distribution is normal, not tailed.
(i) Behavioral data:
Software requirements: R v4.0.5, RStan v2.21.2, rethinking package v2.13, rstanarm package v2.21.1, bayesplot v1.8.0, ggplot2 v3.4.0, lme4 v.1.1.30, MuMIn, afex.
Installation guides can be found on: https://cran.r-project.org/, https://mc-stan.org/users/interfaces/rstan, https://github.com/rmcelreath/rethinking, https://cran.r-project.org/web/packages/rstanarm/index.html, https://mc-stan.org/bayesplot/, https://ggplot2.tidyverse.org/, https://cran.r-project.org/web/packages/lme4/index.html, https://cran.r-project.org/web/packages/MuMIn/index.html, https://cran.r-project.org/web/packages/afex/index.html.
Scripts:
‘Script_Behavior_Bayesian’: Loads behaviour from PD patients (‘HDDM.csv’) and from healthy controls (‘HDDM_HC.csv’), and computes Bayesian hierarchical regression models of reaction times, movement times, peak force, accuracy and regression between reaction times and movement times. It also plots the relevant posteriors.
‘Script_Behavior_NonBayesian’: Conducts the non-Bayesian equivalents of the above mentioned analyses.
(ii) Drift Diffusion Modelling:
Software requirements: Python v3.6, Hierarchical drift diffusion modelling (HDDM v0.8.0), arviz.
Installation guides can be found on: https://www.python.org/downloads/, http://ski.clps.brown.edu/hddm_docs/, https://arviz-devs.github.io/arviz/
Scripts:
‘Scripts_HDDM_Behaviour.py’: Loads behaviour from PD patients (‘HDDM.csv’) and from healthy controls (‘HDDM_HC.csv’), conducts drift diffusion modelling with all possible effects (i), a reduced model with only Instruction effects on decision thresholds (ii) for model convergence checks and posterior predictive checks, and a regression model with single trial movement times as predictor (iii).
(iii) Local field potential data:
Software requirements: Matlab (2019a, requires a software license), FieldTrip v20201126.
Installation guides can be found on: https://matlab.mathworks.com/, https://www.fieldtriptoolbox.org/download/.
Scripts:
‘Script_LFP_FirstLevel.m’: Loads data and applies preprocessing, time-frequency analysis and re-aligning of data using FieldTrip. It uses the custom-written functions ‘ScriptFunction_MakeMontage_AllBipolar’ (which creates a bipolar montage from the monopolar data) and ‘ScriptFunction_EpochData’ (which epochs the continuous data aligned to the moving dots cue and movement onset). The epoched spectra of contralateral and ipsilateral subthalamic nucleus are saved.
‘Script_LFP_SecondLevel_TrialAverages.m’: Loads the spectra from first level analysis and plots the grand average as well as group-averaged theta, beta and gamma traces for all trials, Accuracy trials and Speed trials, for cue- and movement-aligned data.
‘Script_LFP_SecondLevel_SingleTrials.m’: Loads the spectra from first level analysis and computes single trial values of cue-aligned theta, cue-aligned beta, movement-aligned beta and movement-aligned gamma based on time windows of interest. It saves the single trial values together with behavioural data (accuracy, reaction times and movement times) separately for cue-aligned beta, movement-aligned beta and movement-aligned gamma in one csv file and for cue-aligned theta in a separate csv file. These files are used for regression analyses and HDDM. It also saves a csv file with reaction times < 0.4 s excluded for control analyses of cue-aligned beta.
(iv) Regression between local field potential and behavioural data:
Software requirements: R v4.0.5, RStan v2.21.2, rethinking package v2.13, rstanarm package v2.21.1, bayesplot v1.8.0, ggplot2 v3.4.0, lme4 v.1.1.30, MuMIn, afex.
Installation guides: see above.
Scripts:
‘Script_LFP_Bayesian’: Loads behaviour and LFP data from PD patients (‘HDDM_LFP.csv’), and computes Bayesian hierarchical regression models using reaction times and movement times as dependent variables including putative post-hoc tests. It also plots the relevant posteriors. For theta a separate file is loaded (‘HDDM_LFP_Theta.csv’).
‘Script_LFP_NonBayesian’: Conducts the non-Bayesian equivalents of the above mentioned analyses.
(v) Drift Diffusion modelling with local field potential data:
Software requirements: Python v3.6, Hierarchical drift diffusion modelling (HDDM v0.8.0), arviz.
Installation guides: see above.
Scripts:
‘Scripts_HDDM_LFP.py’: Loads behaviour and LFP data from PD patients (‘HDDM_LFP.csv’) and conducts drift diffusion regression modelling with cue-aligned beta and cue- aligned theta (for theta the file ‘HDDM_LFP_Theta.csv’ is loaded).
(vi) DBS effects on behaviour:
Software requirements: Matlab (2019a, requires a software license)
Installation guides: see above.
Scripts:
‘Script_StimulationTime’ loads a file with the stimulation trace during the task, calls the function ‘ScriptFunction_DownsampleBinaryRemoveRamp’ (which downsamples the data to 1000Hz, makes stimulation binary (1 for ON, 0 for OFF) and removes the ramping so that only stimulation at effective intensities counts as stimulation) and loads the behavioural data (reaction time and movement time). It then calls the functions ‘ScriptFunction_WindowedStim’ (which computes for each trial whether or not stimulation was given in any 100 ms moving windows for cue- and movement aligned data), ‘ScriptFunction_WindowedRT’ (which computes reaction times for windows in which stimulation was applied vs. was not applied, also for Accuracy and Speed trials) and ‘ScriptFunction_WindowedMT’ (which computes the analogous measures for movement times). The results are saved separately for reaction times and movement times.
‘Script_StimulationEffects’ loads this data, plots effects of stimulation on movement times and reaction times and provides statistics using cluster-based permutation tests (‘ScriptFunction_PermTest’). It also saves single trial behavioural data with a column stating whether DBS was applied in the critical time window (which is used for the DBS effects on local field potentials analysis).
(vii) DBS effects on local field potentials:
Software requirements: Matlab (2019a, requires a software license), FieldTrip v20201126.
Installation guides: see above.
Scripts:
‘Script_LFP_FirstLevel_Stim.m’: Loads data and applies preprocessing, time-frequency analysis and re-aligning of data using FieldTrip analogously to the script described under (iii) except that it also detrends and demeans the data, applies a low-pass filter at 100 Hz and excludes noisy data points, which are then interpolated. The epoched spectra of contralateral and ipsilateral subthalamic nucleus are saved.
‘Script_LFP_SecondLevel_Stim.m’: Loads data from the previous analysis, loads single trial data with info whether DBS was applied at critical time windows and plots these beta traces together with beta power off stimulation. To exclude that the DBS effect on reaction times might explain any changes, single trial beta power is capped at the single trial reaction times so that no movements fall into the plotted beta power.
Of note, the scripts for unilateral DBS are identical to the scripts described in (vi) and (vii) with the exception that stimulation is analysed for responses with the contralateral and ipsilateral hand.
We welcome researchers wishing to reuse our data to contact the creators of datasets. If you are unfamiliar with analysing the type of data we are sharing, have questions about the acquisition methodology, need additional help understanding a file format, or are interested in collaborating with us, please get in touch via email. Our current members have email addresses on our main site. The corresponding author of an associated publication, or the first or last creator of the dataset are likely to be able to assist, but in case of uncertainty on who to contact, email Ben Micklem, Research Support Manager at the MRC BNDU.
Creative Commons Attribution-ShareAlike 4.0 International (CC BY-SA 4.0)
This is a human-readable summary of (and not a substitute for) the licence.
You are free to:
Share — copy and redistribute the material in any medium or format
Adapt — remix, transform, and build upon the material for any purpose, even commercially.
This licence is acceptable for Free Cultural Works. The licensor cannot revoke these freedoms as long as you follow the license terms. Under the following terms:
Attribution — You must give appropriate credit, provide a link to the license, and indicate if changes were made. You may do so in any reasonable manner, but not in any way that suggests the licensor endorses you or your use.
ShareAlike — If you remix, transform, or build upon the material, you must distribute your contributions under the same licence as the original.
No additional restrictions — You may not apply legal terms or technological measures that legally restrict others from doing anything the licence permits.